Single-cell RNA sequencing (scRNA-seq) has revolutionized our understanding of cellular diversity and function, offering unprecedented insights into complex biological systems. However, the manual annotation of cell types from scRNA-seq data is a laborious and time-consuming process, often requiring months of expert analysis. In recent years, automated methods have emerged to streamline this process, but their accuracy and reliability have been a subject of debate. Now, a groundbreaking study from Columbia University’s Mailman School of Public Health has demonstrated the remarkable potential of GPT-4, a cutting-edge language model, to accurately annotate cell types in scRNA-seq data, rivaling and sometimes surpassing human expert annotation methods.
The Rise of GPT-4: GPT-4, a state-of-the-art language model developed for speech understanding and generation, has garnered attention for its impressive capabilities in various domains. Leveraging its vast knowledge base and sophisticated algorithms, researchers at Columbia University sought to harness the power of GPT-4 for cell type annotation in scRNA-seq data. The study, published in Nature Methods, showcases the remarkable accuracy and efficiency of GPT-4 in deciphering complex cellular landscapes across diverse tissues and species.
Unveiling GPTCelltype: Central to the study is the development of GPTCelltype, an innovative R software package tailored for automated cell type annotation using GPT-4. This user-friendly tool empowers researchers to streamline the annotation process, reducing the reliance on manual expertise and expediting scientific discovery. With GPTCelltype, the labor-intensive task of comparing genes across cell clusters can now be automated, revolutionizing the way researchers interpret and analyze scRNA-seq data.
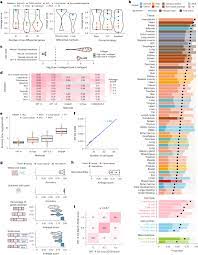
Assessing GPT-4’s Performance: The researchers conducted a comprehensive evaluation of GPT-4’s performance across ten datasets spanning multiple species and tissue types, including both normal and cancer samples. Through meticulous analysis, they demonstrated that GPT-4 consistently produced cell type annotations that closely aligned with those generated by human experts. Notably, GPT-4 exhibited superior accuracy in identifying stromal or connective tissue cells, highlighting its potential to outperform traditional manual methods in specific contexts.
Robustness and Reliability: Beyond its accuracy, GPT-4 showcased remarkable robustness in complex real-world scenarios, accurately distinguishing between pure and mixed cell types with impressive precision. Moreover, GPT-4 demonstrated a high level of consistency in reproducing annotations across different datasets, underscoring its reliability as a tool for scRNA-seq analysis. While acknowledging the limitations of GPT-4 and the challenges of verifying its training procedures, the researchers remain optimistic about its potential to revolutionize cell type annotation in the field of single-cell genomics.
Future Directions and Implications: the integration of GPT-4 into routine scRNA-seq analysis workflows holds tremendous promise for advancing our understanding of cellular biology and disease mechanisms. By democratizing access to advanced annotation tools, GPTCelltype has the potential to accelerate research progress and facilitate collaboration across disciplines. As researchers continue to refine and optimize GPT-4 for specific applications, the era of semi- or fully automated cell type annotation heralds a new chapter in the quest to unravel the complexities of the human genome.
In the age of big data and artificial intelligence, GPT-4 emerges as a transformative force in the realm of single-cell genomics. Its ability to accurately annotate cell types in scRNA-seq data with high consistency and efficiency marks a significant milestone in biomedical research. As scientists harness the power of GPT-4 to unravel the mysteries of cellular diversity, the boundaries of discovery are poised to expand, unlocking new insights into health and disease. With GPT-4 leading the way, the future of single-cell genomics shines bright with possibilities.